Abstract
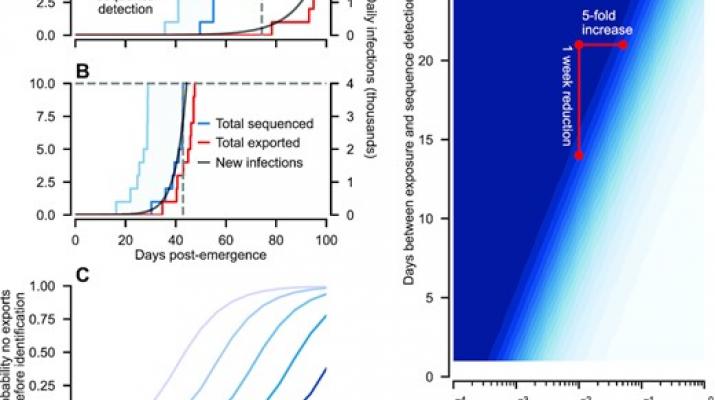
Throughout the COVID-19 pandemic, control of transmission has been repeatedly thwarted by the emergence of variants of concern (VOC) and their geographic spread. Key questions remain regarding effective means of minimizing the impact of VOC, in particular the feasibility of containing them at source, in light of global interconnectedness. By analysing a stochastic transmission model of COVID-19, we identify the appropriate monitoring requirements that make containment at source feasible. Specifically, precise risk assessment informed primarily by epidemiological indicators (e.g. accumulated hospitalization or mortality reports), is unlikely prior to VOC escape. Consequently, decision makers will need to make containment decisions without confident severity estimates. In contrast, successfully identifying and containing variants via genomic surveillance is realistic, provided sequence processing and dissemination is prompt.